The omicron variant appears to have arisen in Africa, perhaps Botswana, and was first detected by the viral sequencing efforts of neighboring South Africa in November of 2021. The changes in Spike protein, relative to other variants, have concerned virologists and public health personnel. The Spike of omicron has 30 amino acid changes, one insertion of three amino acids and three small deletions when compared to the original Wuhan strain. The net effect of the insertions and deletions is the loss of three amino acids relative to the Wuhan strain.
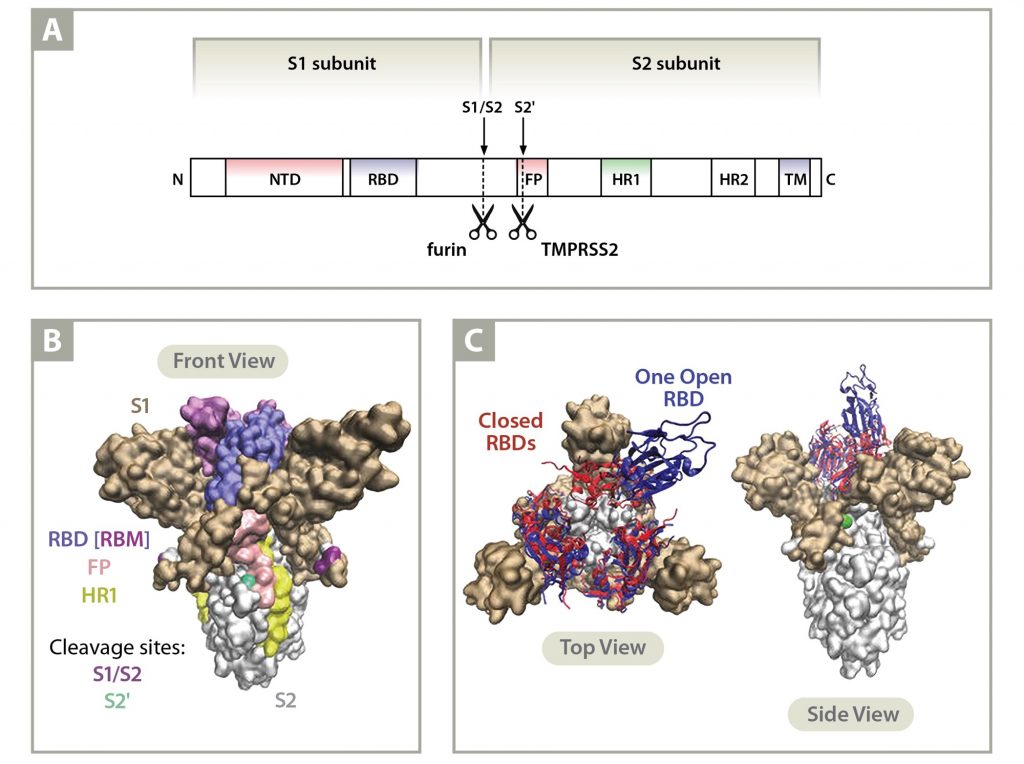
Spike is the best studied of all the proteins produced by the SARS-CoV-2 virus. It largely, but not solely, governs the host immune response to the virus. Spike is responsible for both the adherence of the virus to host cells and the invasion of the virus into cells. Those two features are critical for viral transmission. All the vaccines that are currently available worldwide target Spike.
Amino acid substitutions in Spike can affect both the transmissibility of the virus and how readily the virus avoids neutralizing antibodies. The variety and plenitude of neutralizing antibodies in the host appears to correlate with how easily the host can avoid, or rapidly shake off, infection with the virus. The specific mutations in the Spike omicron variant give reason for concern that it could be both more transmissible and more elusive. This assessment requires the caveat that the combination of these mutations can result in unforeseen effects in the structure of Spike that could vitiate some of these potentially concerning features.
Increased Transmissibility of Omicron Relative to Other Variants is Possible
Until delta arose, alpha was the reigning champion of transmissibility worldwide. SARS-CoV-2, like MERS-CoV, but unlike SARS-CoV, has a polybasic cleavage site in the Spike protein at the S1/S2 junction (residues 681-685). The alpha variant of SARS-CoV-2 experienced a histidine for proline substitution in the polybasic cleavage site (Pro681His) which increased the likelihood that a host protease would cut Spike, thus better preparing it for fusion with a cell. Elimination of proline removed a glycosylation site found in the ancestral Wuhan strain. The attached carbohydrate chain overhangs the cleavage site, inhibiting the approach of proteases. The alteration to histidine also had the advantage, for the virus, of increasing the number of positive charges at physiological pH at that site, which is necessary for host furin-like proteases to cut. Cleavage of Spike at the S1/S2 junction, ideally as it is leaving the incubating cell, increases the chance that the newly made virus will be able to fuse quickly with the next host membrane that it binds(1). The alternative is that the virus sits on the cell membrane in a race to see what happens first, either a host surface protease like TMPRSS2 cuts near the S1/S2 site or the host cell internalizes the virus via the endosomal pathway. The latter increases the chance that a viral component is recognized by host innate immune receptors to instigate a protective cellular response to the detriment of viral infection.
The delta variant was more transmissible than alpha partially because it has a Pro681Arg mutation. Unlike histidine, which displays a positive charge at mildly basic pH, arginine is positively charged under all conditions. This is thought to give delta an advantage over alpha because it improves the chance that S1/S2 site is cut before Spike in the newly made virus leaves the incubating cell(2).
The omicron variant has the same polybasic cleavage site alteration as the alpha variant (Pro681His), so it might be assumed to be less infectious than Delta. But it has an additional alteration which is likely to affect the polybasic cleavage site: Asn679Lys. This adds another positive charge just two residues away from the cleavage site, potentially extending it. How exactly this alters the probability of cleavage awaits investigation, but adding positive charges to the region would be expected to increase the likelihood the S1/S2 cleavage occurs early in the viral life cycle. And this would be expected to increase transmissibility.
Other mutations in the Spike protein of omicron that are likely to increase transmissibility include Ser477Asn and Asn501Tyr. The former enhances the affinity of Spike binding to the human ACE2 receptor by 1.5-fold(3). The latter mutation is common to all variants of concern except delta. N501Y enhances association with the ACE2 receptor by 10-fold compared to the Wuhan strain spike protein(4, 5). For these reasons of structural biology, it is possible omicron could be more transmissible than delta.
Increased Resistance to Existing Neutralizing Antibodies
We have not yet seen a variant that can “break” the neutralizing antibody response induced by vaccination. Vaccination typically results in scores of neutralizing antibody types and no more than a handful have been successfully resisted by any particular variant. This is to say that the typical neutralizing antibody response to vaccination is robust. However, the Spike of omicron features more concerning mutations for the antibody response than any other SARS-CoV-2 that has yet been documented. The most concerning mutations likely to result in an increase in resistance to neutralizing antibodies, and for which there are examples in the literature, are detailed in the table below:
| Omicron Spike Mutation | Reason for Concern |
| Deletion at Val143-Tyr144-Tyr145 | The Y144-Y145 deletion is found in the Spike alpha variant. The resistance of alpha to most monoclonal antibodies in the N-terminal domain (NTD) is largely conferred by this deletion. Mutations in this region abolish the binding of monoclonal antibody 4A8(6, 7). |
| Asn440Lys | The N440K mutation was observed in a virus isolated in India associated with patient re-infection and described as an “immune escape variant”(8). N440K emerged under selection pressure against the human monoclonal antibody C135(9). |
| Gly446Ser | Other mutations at this position in different variants have conferred escape from multiple antibodies(10, 11). |
| Ser477Asn | This S477N mutation in the spike protein is resistant to neutralization by multiple monoclonal antibodies and resulted in a degree of resistance across the entire panel of antibodies(11). |
| Thr478Lys | The T478K mutation is also seen in Delta(12). Other alterations at this position have provided resistance to neutralizing antibodies(11). |
| Glu484Ala | The E484A mutation confers resistance to monoclonal antibody 2B04 and 1B07(11). |
| Gln493Lys | Q493R/K mutations escape neutralization by the monoclonal antibody cocktail LY-CoV555 + LY-CoV016(13). Mutations at Q493 are generally well tolerated with respect to ACE2 binding and expression of the receptor binding domain (RBD) of spike. Q493K confers a greater than two log reduction in IC50 for the REGN10989/10934 pair of monoclonal antibodies compared to the unmutated spike protein(14). |
| Gln498Arg | The Q498R mutation confers escape against the COV2-2499 antibody(10). |
| Asp796Tyr | The substitution of His at this position in Spike modestly decreases sensitivity to convalescent plasma(15). |
Conclusion
The mutations in the Spike protein of the omicron variant of SARS-CoV-2 are likely to result in a more transmissible variant and one that can evade neutralizing antibodies induced by vaccination better than previously seen variants. This is not to suggest that vaccination is likely to be defeated by the omicron variant, only that the protection conferred by vaccination is likely to be shakier than with previous variants of concern. The activated immune system is a potent defender, however, with many weapons to fight infection that are not altogether dependent upon the antibody response.
For more information:

Gene Godbold, PhD
Senior Scientist
Gene Godbold, a Senior Scientist at Signature Science, has been annotating SARS-CoV-2 variants for a US CDC-supported project since June of 2021. The project is led by Professor Todd Treangen of Rice University and is organizing access to SARS-CoV-2 sequences. Dr. Godbold has been analyzing infectious biothreats since 2005 as a government contractor. Supporting IARPA’s Fun GCAT program, he has delivered leading-edge work in biological threat determination, developing criteria to categorize biothreat sequences and curating thousands of threat sequences from bacteria, viruses, fungi and other parasites from the professional literature. Dr. Godbold is conversant in the mechanisms of virulence factors from hundreds of infectious agents, particularly at the microbial/host immune interface. His areas of expertise include cellular and molecular biology, biochemistry, microbiology, immunology, and infectious diseases.
Citations
- Zhang L, Mann M, Syed Z, Reynolds HM, Tian E, Samara NL, Zeldin DC, Tabak LA, Ten Hagen KG. 2021. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. bioRxiv 2021.02.05.429982.
- Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, Widen SG, An Z, Weaver SC, Menachery VD, Xie X, Shi P-Y. 2021. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv 2021.08.12.456173.
- Barton MI, MacGowan SA, Kutuzov MA, Dushek O, Barton GJ, van der Merwe PA. 2021. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife 10:e70658.
- Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, Oliveira TY, Yang Z, Abernathy ME, Huey-Tubman KE, Hurley A, Turroja M, West KA, Gordon K, Millard KG, Ramos V, Da Silva J, Xu J, Colbert RA, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Gazumyan A, Caskey M, Bjorkman PJ, Casellas R, Hatziioannou T, Bieniasz PD, Nussenzweig MC. 2021. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592:616–622.
- Luan B, Wang H, Huynh T. 2021. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett 595:1454–1461.
- Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593:130–135.
- McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, Duprex WP. 2021. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371:1139–1142.
- Rani PR, Imran M, Lakshmi JV, Jolly B, Jain A, Surekha A, Senthivel V, Chandrasekhar P, Divakar MK, Srinivasulu D, Bhoyar RC, Vanaja PR, Scaria V, Sivasubbu S. 2021. Symptomatic reinfection of SARS-CoV-2 with spike protein variant N440K associated with immune escape. J Med Virol 93:4163–4165.
- Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann H-H, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9:e61312.
- Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE, Bloom JD. 2021. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29:44-57.e9.
- Liu Z, VanBlargan LA, Bloyet L-M, Rothlauf PW, Chen RE, Stumpf S, Zhao H, Errico JM, Theel ES, Liebeskind MJ, Alford B, Buchser WJ, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ. 2021. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29:477-488.e4.
- Arora P, Rocha C, Kempf A, Nehlmeier I, Graichen L, Winkler MS, Lier M, Schulz S, Jäck H-M, Cossmann A, Stankov MV, Behrens GMN, Pöhlmann S, Hoffmann M. 2021. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell Mol Immunol https://doi.org/10.1038/s41423-021-00779-5.
- Starr TN, Greaney AJ, Dingens AS, Bloom JD. 2021. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. bioRxiv 2021.02.17.431683.
- Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. 2020. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369:1014–1018.
- Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, Roberts DJ, Chandra A, Temperton N, CITIID-NIHR BioResource COVID-19 Collaboration, COVID-19 Genomics UK (COG-UK) Consortium, Sharrocks K, Blane E, Modis Y, Leigh KE, Briggs JAG, van Gils MJ, Smith KGC, Bradley JR, Smith C, Doffinger R, Ceron-Gutierrez L, Barcenas-Morales G, Pollock DD, Goldstein RA, Smielewska A, Skittrall JP, Gouliouris T, Goodfellow IG, Gkrania-Klotsas E, Illingworth CJR, McCoy LE, Gupta RK. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592:277–282.

