
Genomics Specialist
Contact Krista
The prevalence of healthcare-acquired infections (HAI) and rising levels of antimicrobial resistance places significant economic and public health burdens on modern healthcare systems. A group of highly drug resistant pathogens known as the ESKAPE pathogens, along with C. difficile, are the leading causes of HAIs. Interactions between patients, healthcare workers, and environmental conditions impact disease transmission.
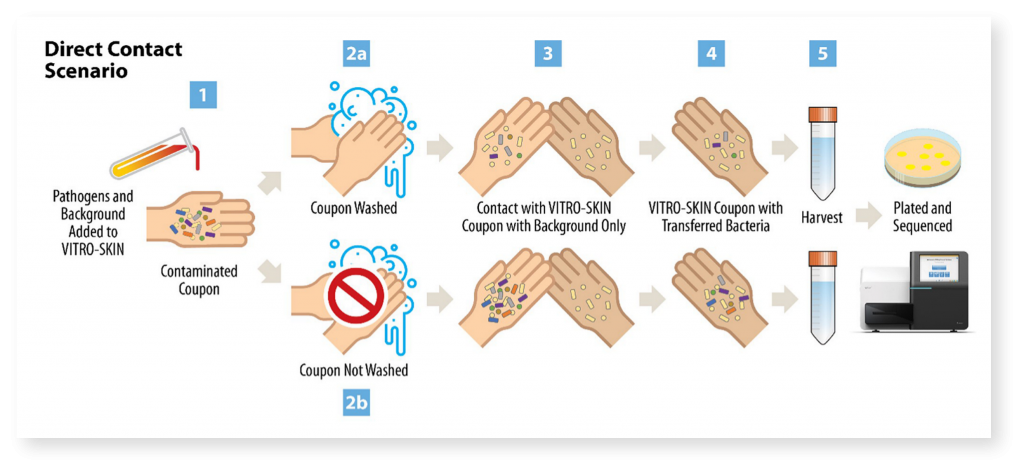
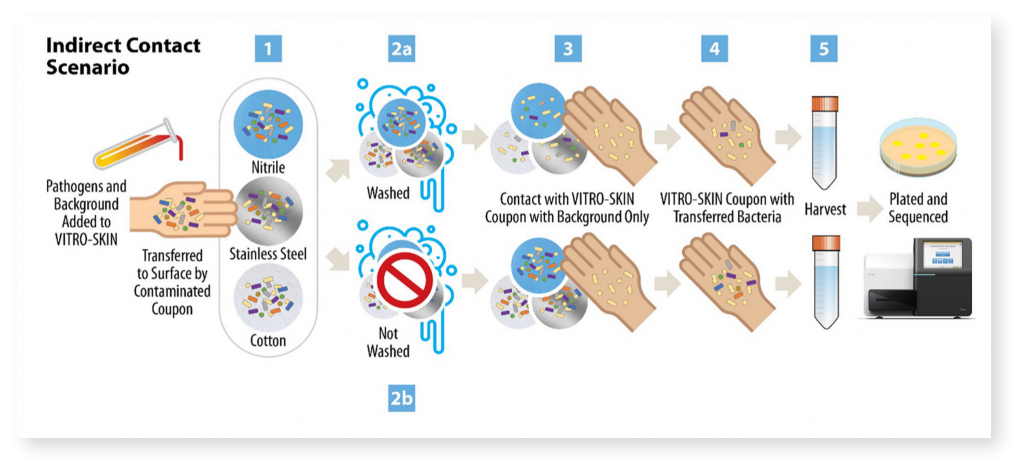
With the goal of contributing to improvements in pathogen transmission modeling in healthcare settings, Signature Science conducted research supported by the Centers of Disease Control and Prevention’s investments to combat antibiotic resistance under award number (200-2018-75D30118C02922). Study objectives were to evaluate how healthcare-associated pathogen transfer rates change under a variety of simulated conditions, compare traditional culturing methods with the culture-independent methods of shotgun metagenomics and metatranscriptomics for pathogen detection and characterization, assess how pathogen abundance impacts detection of virulence and antimicrobial resistance genes, and identify potential signatures of pathogen viability.
Experimental Design
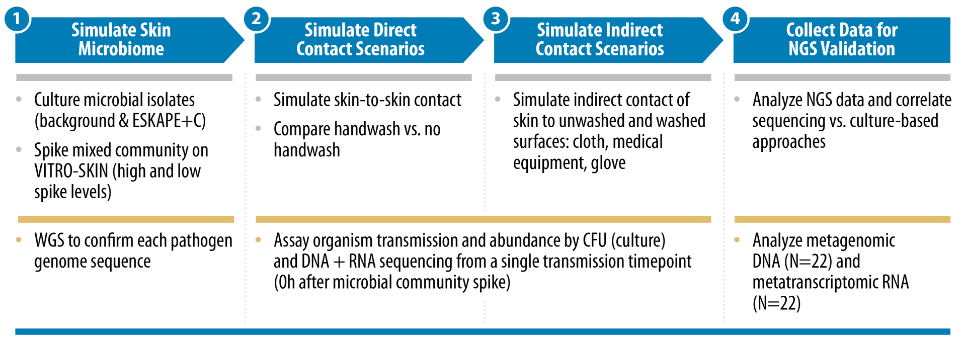
Evaluating Pathogen Transfer Rates Under Various Conditions
Publications
Data and Analysis
For this project’s sequencing data, visit:
For intermediate analyses and results from tools, visit:
Want more information about this project?
