
Molecular Biologist
Contact Anthony
The SARS-CoV-2 pandemic illustrated that RNA viruses pose a significant global biothreat. Unfortunately, the detection of RNA viruses can be a major challenge, especially for novel or emerging infectious disease viruses that lack well-established targeted detection assays.
Universal or ‘target agnostic’ RNA sequencing and related computational analysis has the potential to overcome these challenges to enable the detection of any virus or pathogen present in a sample. Sequencing approaches continue to advance in speed and accuracy with lower associated costs. Nanopore sequencing highlights this trend by combining real-time sequence analysis with relatively inexpensive sequencing reagents.
Validated Target Agnostic Sequencing
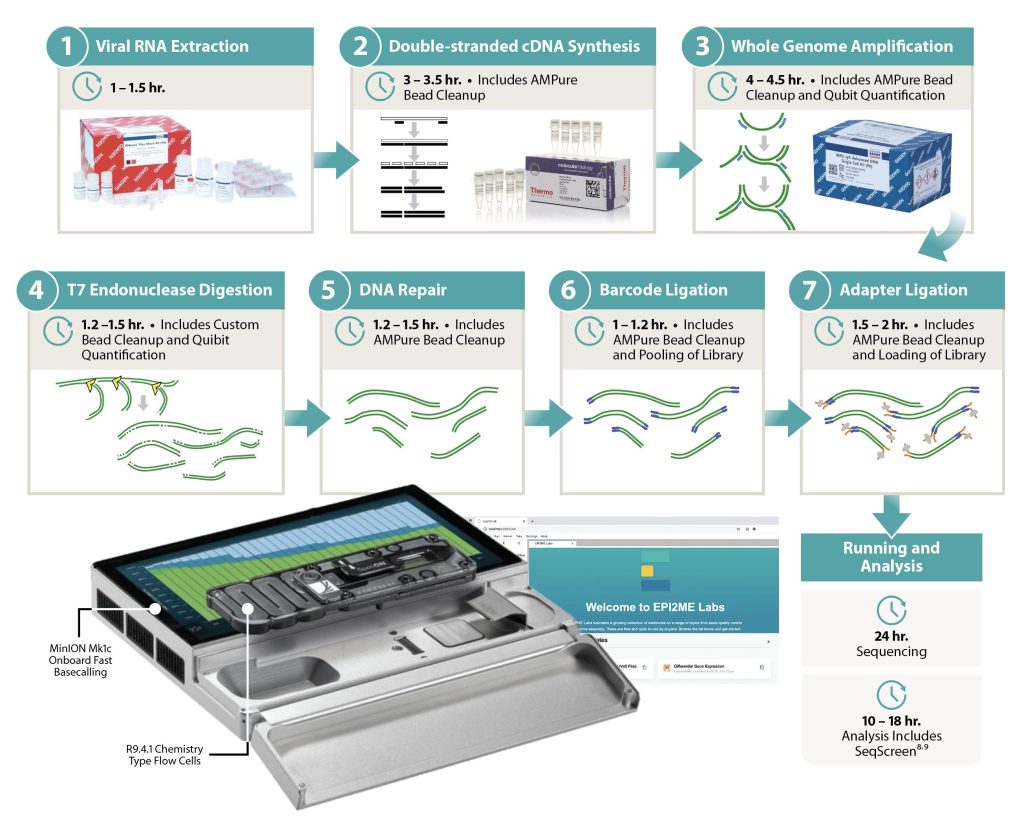

With funding from the Centers for Disease Control and Prevention (CDC), Signature Science evaluated and optimized a suite of standardized protocols using the Oxford Nanopore Technologies handheld-sized MinION sequencing device to rapidly and accurately detect RNA viruses categorized as biothreats. Our method includes a custom bioinformatics and analysis reporting pipeline, SigSciDx, which performs accurate determination of biothreat presence. Assay limits of detection rival that of common immunoassays, providing sufficient sensitivity for clinical settings. Our results demonstrate that clinical metagenomic sequencing can serve as a single universal assay requiring no previous knowledge of the presence of pathogens, including novel, emerging, or rare viruses.
With continued funding from the CDC, Signature Science has validated a target agnostic sequencing assay which includes SigSciDx bioinformatic analysis for the detection of RNA virus pathogens in common clinical sample types. We have also prepared for the future transition of this method to public health laboratories through the development of an implementation plan and laboratory training course. We have already completed a pilot implementation to a public health laboratory in the CDC Laboratory Response Network within a Biosafety Level 3 laboratory.
Want more information about Universal Detection for Biothreats and Emerging Diseases?
FUNDING ACKNOWLEDGEMENT: This research was funded by the Department of Health and Human Services (HHS), Centers for Disease Control and Prevention (CDC) under award numbers 75D30121C12250 and 75D30122C15359. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the HHS, CDC, or the US Government.
